Publications
Below are selected works highlighting our most significant research contributions to date. For a complete list of academic publications from our lab, see Google Scholar.
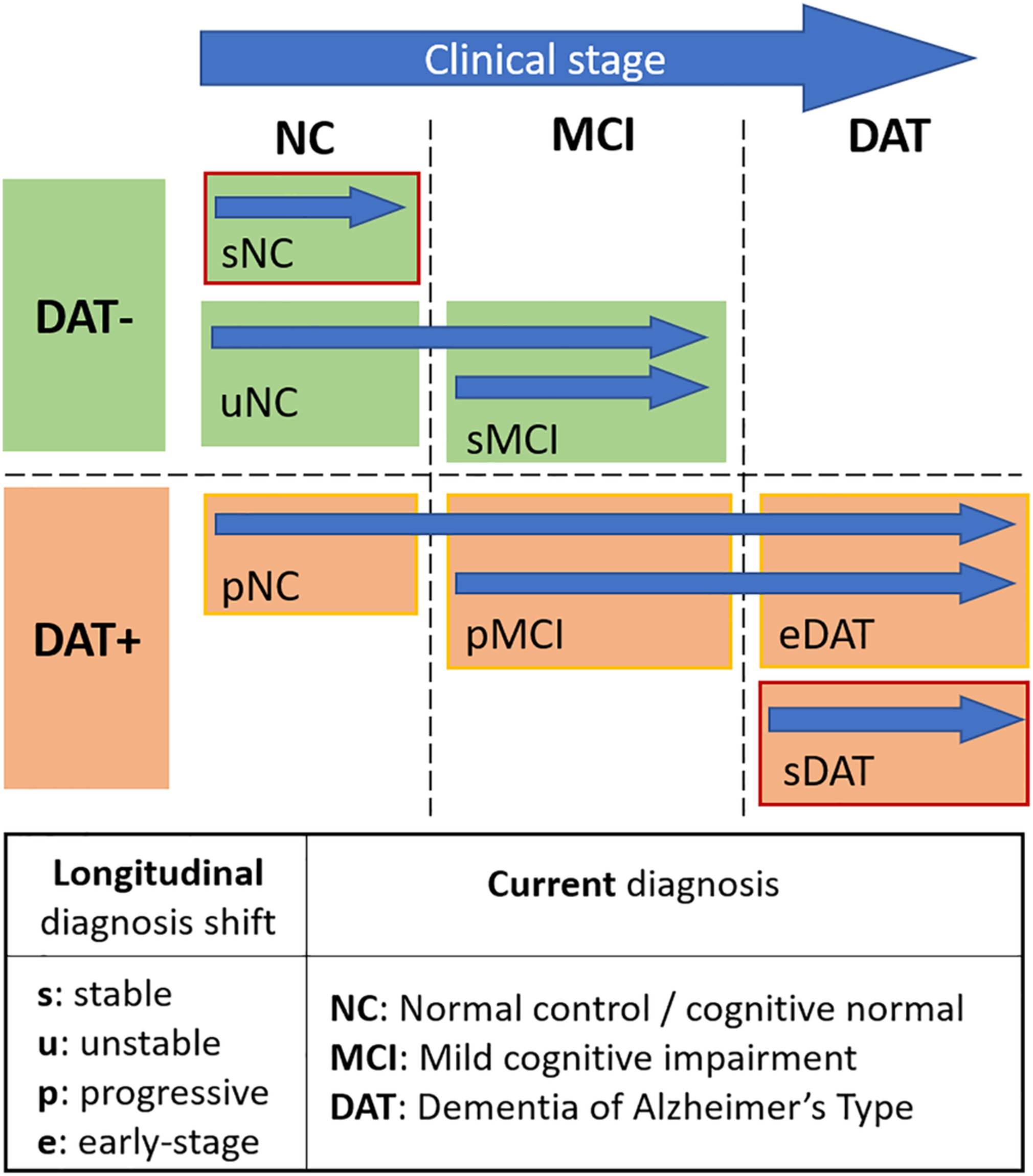
A novel biomarker using structural MRI volume-based features for dementia of Alzheimer’s type (DAT) group. Classification performance on stable versus progressive mild cognitive impairment (MCI) groups achieved an AUC of 0.73 for time to conversion (TTC) of up to 7 years.
Popuri K., Ma D., Wang L., Beg M. F.
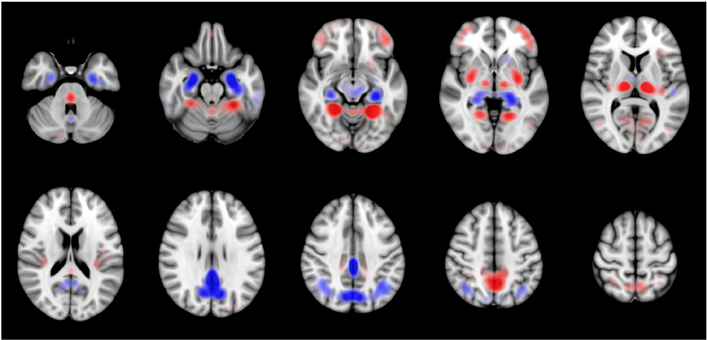
This work proposes a 3D convolutional neural network with residual connections that generates a probability score useful for interpreting the FDG-PET images along the continuum of AD. Classification performance of 0.811 AUC is achieved in the task of predicting conversion of mild cognitive impairment to DAT for conversion time of 0-3 years. The saliency and class activation maps, which highlight the regions of the brain that are most important to the classification task, implicate many known regions affected by DAT including the posterior cingulate cortex, precuneus, and hippocampus.
Yee E., Popuri K., Beg M. F.

A novel FDG-PET biomarker indicating probability of dementia of Alzheimer’s type (DAT) diagnosis. Comprehensive biomarker evaluation using 2984 ADNI images achieved a good validation AUC of 0.78.
Popuri K., Balachandar R., Alpert K., Lu D., Bhalla M., Mackenzie I.R., Hsiung R.G-Y., Wang L., Beg M.F.

A deep learning based classification model for discriminating individuals with AD utilizing a multimodal and multiscale deep neural network. Our method delivers 82.4% accuracy in identifying the individuals with mild cognitive impairment (MCI) who will convert to AD at 3 years prior to conversion (86.4% combined accuracy for conversion within 1-3 years), a 94.23% sensitivity in classifying individuals with clinical diagnosis of probable AD, and a 86.3% specificity in classifying non-demented controls improving upon results in published literature.
Lu D., Popuri K., Ding G.W., Balachandar R., Beg M.F.
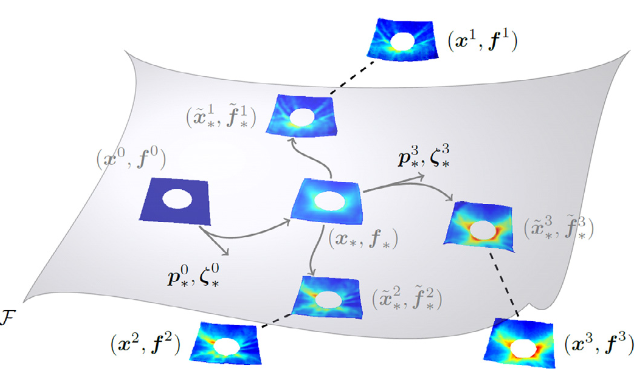
We propose a novel approach for quantitative shape variability analysis in retinal optical coherence tomography images using the functional shape (fshape) framework. To demonstrate the clinical relevance and application of the framework, we generated atlases of the inner layer surface and layer thickness of the Retinal Nerve Fiber Layer (RNFL) of glaucomatous and normal subjects, visualizing detailed spatial pattern of RNFL loss in glaucoma.
Lee S., Charon N., Charlier B., Popuri K., Lebed E., Sarunic M.V., Trouve A., Beg M.F.

The first ever fully automated framework for segmentation of muscle and fat tissues from CT images to estimate body composition. We developed a novel finite element method (FEM) deformable model that incorporates a priori shape information via a statistical deformation model (SDM) within the template-based segmentation framework. The proposed method was validated on 1000 abdominal and 530 thoracic CT images and we obtained very good segmentation results with Jaccard scores in excess of 90% for both the muscle and fat regions.
Popuri K., Cobzas D., Esfandiari N., Baracos V., Jagersand M.
IEEE Transactions on Medical Imaging, 2016
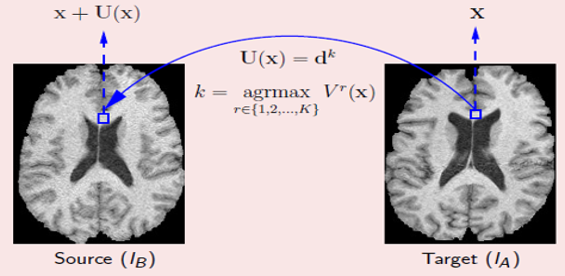
We present a novel variational formulation of discrete deformable registration as the minimization of a convex energy functional that involves diffusion regularization. A computationally efficient solution using the finite element method (FEM) method has been proposed to solve the variational minimization problem. Our proposed method obtained competitive results when compared with 14 other deformable registration methods on the CUMC12 MRI dataset.
Popuri K., Cobzas D., Jagersand M.

One of the earliest methods proposing a fully automated method for brain tumor segmentation. The available MRI modalities (T1, T1c, T2) are used to devise a texture classification based approach within a 3D variational segmentation framework. The segmentation performance was evaluated on 15 real MRI scans with Jaccard score of 58%.
Popuri K., Cobzas D., Murtha A., Jagersand M.
International Journal of Computer Assisted Radiology and Surgery, 2012